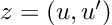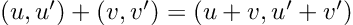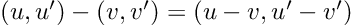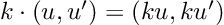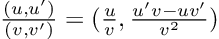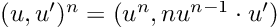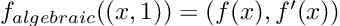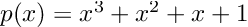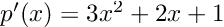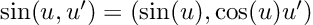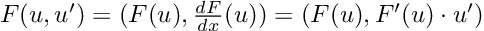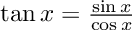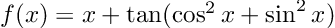Re-posted from: http://www.stochasticlifestyle.com/chatgpt-performs-better-on-julia-than-python-and-r-for-large-language-model-llm-code-generation-why/
Machine learning is all about examples. The more data you have, the better it should perform, right? With the rise of ChatGPT and Large Language Models (LLMs) as a code helping tool, it was thus just an assumption that the most popular languages like Python would likely be the best for LLMs. But because of the increased productivity, I tend to use a lot of Julia, a language with an estimated user-base of around a million programmers. For this reason, people have often asked me how it fairs with ChatGPT, Github Copilot, etc., and so I checked out those pieces and… was stunned. It’s really good. It seemed better than Python actually?
The data is in: Julia does well with ChatGPT
This question was recently put to the test by a researcher named Alessio Buscemi in A Comparative Study of Code Generation using ChatGPT 3.5 across 10 Programming Languages. Basically, he setup a bunch of example problems and asked ChatGPT for the code, executed it, and documented the results. The main result of the paper is the following:
or in the words of the author:
“Overall, 1833 runs, or 45.8% of the total number, lead to executable code. However, this percentage varies greatly according to the tested language. ChatGTP performs the best on Julia, with a 81.5% of generated code being successfully executed, and performs the worst on C++, with only 7.3% of the executions being successful. Specifically, the model seems to perform better on high-level dynamically typed languages (Javascript, Julia, Perl, Python, R, Ruby, Smalltalk) rather than lower level statically typed languages (C, C++, Go).”
That is right, of all languages, “ChatGTP performs the best on Julia”. In fact, ChatGPT generally only performs well on slow languages: Julia is the only fast language that it did well on. The author went on to do a podcast at DataSkeptic where at 25:20 this is addressed where he was unable to answer why ChatGPT was able to make him more successful in Julia than Python, even though he himself only had used Python before.
Is it unexpected that Julia would outperform Python in generated code execution and correctness?
But the real question is, is this really unexpected? I don’t think so, this aligns with what I have seen. I am an educator (currently at MIT researching/teaching in machine learning) who has ran many workshops over the last 10 years, with the languages mainly being Julia, Python, MATLAB, R, and C. I also recently have been a power-user of this translation because I recently have been updating the bindings and documentation of diffeqpy, a fast differential equation solver package in Python which uses Julia as the backend. This has been requiring a lot of translating Julia DifferentialEquations.jl code into a Python format for diffeqpy, and thus there was some ChatGPT involved.
[A quick intro to the “why of diffeqpy” is that we recently had a peer reviewed study demonstrating the generated GPU kernels from Julia for ODE solvers were about 20x-100x faster than the leading PyTorch and Jax libraries. Given these results, we wanted to create a Julia backend for scientists who use Python and could demonstrate on Google Collab notebooks that even when used through Python with automated language translation, it’s still about 10x faster than Jax. The future is working together: if you happen to be at PyData Eindhoven or PyData Global, come chat with me about building bridges between languages!]
In both of these scenarios, the development of diffeqpy with the help of ChatGPT and in teaching new students Julia vs Python, one of the things that really stuck out was that new developers and AI get caught on the API inconsistencies of Python. “But Python is so Pythonic, it doesn’t have inconsistencies?”… ehh have you tried to teach it? I think it’s an interesting thing in the psyche of a programmer. If you have used a programming language for 10 years, then everything the programming language does feels natural. It’s like an English speaker using the word “do”, a phase by which do-support is very difficult for new language learners, but someone who natively learned English may have never thought twice about how weird the word is.
There is a lot of this “it’s always been like this and therefore it makes sense” in Python. In the workshops, it always got best highlighted when using a cheatsheet which shows Julia vs MATLAB vs Python syntax side-by-side. One of the examples that comes to mind is the syntax for the simplest things, like “let’s create an array of ones, zeros, or random numbers”. What does that look like side-by-side?
The Python form in the middle is undoubtably a bit weird, requires extra packages (“what is np?”), etc. but it’s not “awful”. However, it’s then the rand part that gets students:
And that’s when it all becomes clear. np.zeros((2, 2)) to np.random.rand(2, 2), students always ask “why the ((” and the answer is “well there’s a tuple and …” but no, no matter how much we as educators try to explain it, it’s just an inconsistency. It’s just harder. And it happens over and over in the teaching. Sometimes it’s that the syntax is more complex or opaque:
while in other cases it’s inconsistency, unconventional wording, or something else.
So what happened with ChatGPT? It tripped up on exactly these same points that new learners commonly tripped up on. Common issues I noticed when developing diffeqpy was:
- Results that were “right” but contextually wrong because of standard library inconsistencies or lack of a standard library. For example, “create an array of random numbers in Python” does “import random”, then “random_numbers = [random.randint(1, 100) for _ in range(5)]”. Okay, that’s not wrong, but “obviously” the right thing to do is to create a numpy array in any context where I am using a differential equation solver. But ChatGPT doesn’t know these contextual differences, it does not guess well, and if a user is not already familiar in Python they will get tripped up. The solution is to be more specific “create an array of random numbers in Python with numpy”, i.e. prompt engineering. But notably, this prompt engineering was not required with the same examples in Julia, and is generally a result of Python having a lack of core standardization, i.e. having many different ways to do basic things (numpy vs PyTorch vs standard libraries and methods built into CPython).
- Results that were “close” but tripped up due to extra complexity. “A = coo_matrix(([4, 1],([0, 1], [1, 1])),shape=(2, 2))” is undeniably hard syntax to guess, and ChatGPT messed up quite a bit on examples like these sparse matrix constructions and forced vectorizations. Interestingly, it had some difficulties in translating code to zero-based indexing in a few cases. Mathematical models in texts tend to be written in one-based ($$x_1$$), and giving ChatGPT the extra step of converting one-based to zero-based gave it an extra step which I found tripped it up in a few cases.
- Results that were too inefficient to be used in practice. This is something that tag-teamed the last one. Sometimes the code that would be correct would be a for loop (that would often times have difficulty compiling in numba), but that code would inhibit performance enough that I would ask for a vectorized version, in which case it would create a weird matrix with some syntax errors (which is also not fast after fixing the syntax errors).
- Results that were clearly pulling from “bad data”. More data is not always better. One other interesting point to note was that, in the case of differential equations, it was clear that ChatGPT was relying on data from tutorials and web responses I had written because its examples match my favorite examples (Lotka-Volterra, ROBER, Bruss, etc.) and match a lot of the coding styles I prefer (for reference, I wrote and maintain the differential equation libraries in Julia). In the Python examples I could find some oddities of sometimes inappropriately chosen solvers (stiff vs non-stiff) with some odd statements that you wouldn’t expect an expert to say. What I mean is, the Python code clearly had a larger set of training data, but not everyone in that training data seemed to really know the ins and outs of numerical differential equations to be actually trustworthy data. This likely is one of the major parts impacting the results.
These one of the reasons why people tend to say you need to double check your code when generated by LLMs (or that they aren’t quite ready), it’s because these errors tend to happen (often). However, what I found is that these classes of errors were dramatically increased with Python instead of Julia, and where it tripped up is exactly where I would have expected new students to get tripped up. That’s not to say it does perfect (or well) on Julia, but it clearly did better, and thus after trying hard I tended to only use ChatGPT to convert Python examples to Julia and not Julia examples to Python in the end, simply because the Python generated examples required too much work to fix.
Conclusion
It’s interesting what you can get used to in life. Discomfort can become the norm if it’s what you experience every day. I had a back pain that I thought was just normal because I had gotten older, until I got a new office chair and realized it went away. In the same way, programming languages add their own discomforts to your programming life. You get used to them, but it becomes very obvious where the pain points are when you have to teach somebody new, since they have not become accustomed to the pain. When I first started using Julia, I came because I needed a high level language that generated fast code. When I started moving workshops from MATLAB and Python to Julia, I was shocked at how much easier it was to train newcomers to the language because of the syntactic simplicity. But now when using LLMs, it’s interesting to see how that syntactic simplicity and overall uniformity also reduced errors from AI generated code in precisely the same spots. My “eye check” from the diffeqpy project has now been confirmed by a larger study that indeed Julia works well with LLMs.
Are LLMs ready for doing all things code generation? No. But it does do surprisingly well with Julia, and it will be interesting to watch how that evolves as we get more and more Julia code as training data.
The post ChatGPT performs better on Julia than Python (and R) for Large Language Model (LLM) Code Generation. Why? appeared first on Stochastic Lifestyle.