By: Bradley Setzler
Re-posted from: https://juliaeconomics.com/2014/09/02/selection-bias-corrections-in-julia-part-1/
Selection bias arises when a data sample is not a random draw from the population that it is intended to represent. This is especially problematic when the probability that a particular individual appears in the sample depends on variables that also affect the relationships we wish to study. Selection bias corrections based on models of economic behavior were pioneered by the economist James J. Heckman in his seminal 1979 paper.
For an example of selection bias, suppose we wish to study the effectiveness of a treatment (a new medicine for patients with a particular disease, a preschool curriculum for children facing particular disadvantages, etc.). A random sample is drawn from the population of interest, and the treatment is randomly assigned to a subset of this sample, with the remaining subset serving as the untreated (“control”) group. If the subsets followed instructions, then the resulting data would serve as a random draw from the data generating process that we wish to study.
However, suppose the treatment and control groups do not comply with their assignments. In particular, if only some of the treated benefit from treatment while the others are in fact harmed by treatment, then we might expect the harmed individuals to leave the study. If we accepted the resulting data as a random draw from the data generating process, it would appear that the treatment was much more successful than it actually was; an individual who benefits is more likely to be present in the data than one who does not benefit.
Conversely, if treatment were very beneficial, then some individuals in the control group may find a way to obtain treatment, possibly without our knowledge. The benefits received by the control group would make it appear that the treatment was less beneficial than it actually was; the receipt of treatment is no longer random.
In this tutorial, I present some parameterized examples of selection bias. Then, I present examples of parametric selection bias corrections, evaluating their effectiveness in recovering the data generating processes. Along the way, I demonstrate the use of the GLM package in Julia. A future tutorial demonstrates non-parametric selection bias corrections.
Example 1: Selection on a Normally-Distributed Unobservable
Suppose that we wish to study of the effect of the observable variable 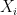 on
on  . The data generating process is given by,
. The data generating process is given by,
 ,
,
where 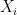 and
and  are independent in the population. Because of this independence condition, the ordinary least squares estimator would be unbiased if the data
are independent in the population. Because of this independence condition, the ordinary least squares estimator would be unbiased if the data  were drawn randomly from the population. However, suppose that the probability that individual
were drawn randomly from the population. However, suppose that the probability that individual  were in the data set
were in the data set  were a function of
were a function of 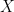 and
and  . For example, suppose that,
. For example, suppose that,
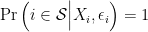 if
if 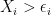 ,
,
and the probability is zero otherwise. This selection rule ensures that, among the individuals in the data (the  in
in 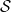 ), the covariance between
), the covariance between 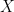 and
and  will be positive, even though the covariance is zero in the population. When
will be positive, even though the covariance is zero in the population. When 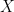 covaries positively with
covaries positively with  , then the OLS estimate of
, then the OLS estimate of 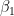 is biased upward, i.e., the OLS estimator converges to a value that is greater than
is biased upward, i.e., the OLS estimator converges to a value that is greater than 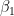 .
.
To see the problem, consider the following simulation of the above process in which 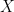 and
and  are drawn as independent standard normal random variables:
are drawn as independent standard normal random variables:
srand(2)
N = 1000
X = randn(N)
epsilon = randn(N)
beta_0 = 0.
beta_1 = 1.
Y = beta_0 + beta_1.*X + epsilon
populationData = DataFrame(Y=Y,X=X,epsilon=epsilon)
selected = X.>epsilon
sampleData = DataFrame(Y=Y[selected],X=X[selected],epsilon=epsilon[selected])
There are 1,000 individuals in the population data, but 492 of them are selected to be included in the sample data. The covariance between X and epsilon in the population data is,
cov(populationData[:X],populationData[:epsilon])
0.00861456704877879
which is approximately zero, but in the sample data, it is,
cov(sampleData[:X],sampleData[:epsilon])
0.32121357192108513
which is approximately 0.32.
Now, we regress 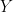 on
on 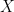 with the two data sets to obtain,
with the two data sets to obtain,
linreg(array(populationData[:X]),array(populationData[:Y]))
2-element Array{Float64,1}:
-0.027204
1.00882
linreg(array(sampleData[:X]),array(sampleData[:Y]))
2-element Array{Float64,1}:
-0.858874
1.4517
where, in Julia 0.3.0, the command array() replaces the old command matrix() in converting a DataFrame into a numerical Array. This simulation demonstrates severe selection bias associated with using the sample data to estimate the data generating process instead of the population data, as the true parameters,  , are not recovered by the sample estimator.
, are not recovered by the sample estimator.
Correction 1: Heckman (1979)
The key insight of Heckman (1979) is that the correlation between 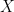 and
and  can be represented as an omitted variable in the OLS moment condition,
can be represented as an omitted variable in the OLS moment condition,
![\mathbb{E}\left[Y_i\Big|X_i,i\in\mathcal{S}\right]=\mathbb{E}\left[\beta_0 + \beta_1X_i+\epsilon_i\Big|X_i,i\in\mathcal{S}\right]=\beta_0 + \beta_1X_i+\mathbb{E}\left[\epsilon_i\Big|X_i,i\in\mathcal{S}\right] \mathbb{E}\left[Y_i\Big|X_i,i\in\mathcal{S}\right]=\mathbb{E}\left[\beta_0 + \beta_1X_i+\epsilon_i\Big|X_i,i\in\mathcal{S}\right]=\beta_0 + \beta_1X_i+\mathbb{E}\left[\epsilon_i\Big|X_i,i\in\mathcal{S}\right]](https://s0.wp.com/latex.php?latex=%5Cmathbb%7BE%7D%5Cleft%5BY_i%5CBig%7CX_i%2Ci%5Cin%5Cmathcal%7BS%7D%5Cright%5D%3D%5Cmathbb%7BE%7D%5Cleft%5B%5Cbeta_0+%2B+%5Cbeta_1X_i%2B%5Cepsilon_i%5CBig%7CX_i%2Ci%5Cin%5Cmathcal%7BS%7D%5Cright%5D%3D%5Cbeta_0+%2B+%5Cbeta_1X_i%2B%5Cmathbb%7BE%7D%5Cleft%5B%5Cepsilon_i%5CBig%7CX_i%2Ci%5Cin%5Cmathcal%7BS%7D%5Cright%5D&bg=ffffff&%23038;fg=333333&%23038;s=0) ,
,
Furthermore, using the conditional density of the standard Normally distributed  ,
,
![\mathbb{E}\left[\epsilon_i\Big|X_i,i\in\mathcal{S}\right]=\mathbb{E}\left[\epsilon_i\Big|X_i,X_i>\epsilon_i\right]=\frac{-\phi\left(X_i\right)}{\Phi\left(X_i\right)} \mathbb{E}\left[\epsilon_i\Big|X_i,i\in\mathcal{S}\right]=\mathbb{E}\left[\epsilon_i\Big|X_i,X_i>\epsilon_i\right]=\frac{-\phi\left(X_i\right)}{\Phi\left(X_i\right)}](https://s0.wp.com/latex.php?latex=%5Cmathbb%7BE%7D%5Cleft%5B%5Cepsilon_i%5CBig%7CX_i%2Ci%5Cin%5Cmathcal%7BS%7D%5Cright%5D%3D%5Cmathbb%7BE%7D%5Cleft%5B%5Cepsilon_i%5CBig%7CX_i%2CX_i%3E%5Cepsilon_i%5Cright%5D%3D%5Cfrac%7B-%5Cphi%5Cleft%28X_i%5Cright%29%7D%7B%5CPhi%5Cleft%28X_i%5Cright%29%7D&bg=ffffff&%23038;fg=333333&%23038;s=0) .
.
which is called the inverse Mills ratio, where  and
and  are the probability and cumulative density functions of the standard normal distribution. As a result, the moment condition that holds in the sample is,
are the probability and cumulative density functions of the standard normal distribution. As a result, the moment condition that holds in the sample is,
![\mathbb{E}\left[Y_i\Big|X_i,i\in\mathcal{S}\right]=\beta_0 + \beta_1X_i+\frac{-\phi\left(X_i\right)}{\Phi\left(X_i\right)} \mathbb{E}\left[Y_i\Big|X_i,i\in\mathcal{S}\right]=\beta_0 + \beta_1X_i+\frac{-\phi\left(X_i\right)}{\Phi\left(X_i\right)}](https://s0.wp.com/latex.php?latex=%5Cmathbb%7BE%7D%5Cleft%5BY_i%5CBig%7CX_i%2Ci%5Cin%5Cmathcal%7BS%7D%5Cright%5D%3D%5Cbeta_0+%2B+%5Cbeta_1X_i%2B%5Cfrac%7B-%5Cphi%5Cleft%28X_i%5Cright%29%7D%7B%5CPhi%5Cleft%28X_i%5Cright%29%7D&bg=ffffff&%23038;fg=333333&%23038;s=0)
Returning to our simulation, the inverse Mills ratio is added to the sample data as,
sampleData[:invMills] = -pdf(Normal(),sampleData[:X])./cdf(Normal(),sampleData[:X])
Then, we run the regression corresponding to the sample moment condition,
linreg(array(sampleData[[:X,:invMills]]),array(sampleData[:Y]))
3-element Array{Float64,1}:
-0.166541
1.05648
0.827454
We see that the estimate for 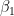 is now 1.056, which is close to the true value of 1, compared to the non-corrected estimate of 1.452 above. Similarly, the estimate for
is now 1.056, which is close to the true value of 1, compared to the non-corrected estimate of 1.452 above. Similarly, the estimate for  has improved from -0.859 to -0.167, when the true value is 0. To see that the Heckman (1979) correction is consistent, we can increase the sample size to
has improved from -0.859 to -0.167, when the true value is 0. To see that the Heckman (1979) correction is consistent, we can increase the sample size to 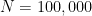 , which yields the estimates,
, which yields the estimates,
linreg(array(sampleData[[:X,:invMills]]),array(sampleData[:Y]))
3-element Array{Float64,1}:
-0.00417033
1.00697
0.991503
which are very close to the true parameter values.
Note that this analysis generalizes to the case in which  contains
contains  variables and the selection rule is,
variables and the selection rule is,
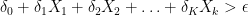 ,
,
which is the case considered by Heckman (1979). The only difference is that the coefficients  must first be estimated by regressing an indicator for
must first be estimated by regressing an indicator for 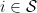 on
on 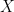 , then using the fitted equation within the inverse Mills ratio. This requires that we observe
, then using the fitted equation within the inverse Mills ratio. This requires that we observe 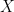 for
for  . Probit regression is covered in a slightly different context below.
. Probit regression is covered in a slightly different context below.
As a matter of terminology, the process of estimating  is called the “first stage”, and estimating
is called the “first stage”, and estimating 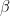 conditional on the estimates of
conditional on the estimates of  is called the “second stage”. When the coefficient on the inverse Mills ratio is positive, it is said that “positive selection” has occurred, with “negative selection” otherwise. Positive selection means that, without the correction, the estimate of
is called the “second stage”. When the coefficient on the inverse Mills ratio is positive, it is said that “positive selection” has occurred, with “negative selection” otherwise. Positive selection means that, without the correction, the estimate of 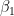 would have been upward-biased, while negative selection results in a downward-biased estimate. Finally, because the selection rule is driven by an unobservable variables
would have been upward-biased, while negative selection results in a downward-biased estimate. Finally, because the selection rule is driven by an unobservable variables  , this is a case of “selection on unobservables”. In the next section we consider a case of “selection on observables”.
, this is a case of “selection on unobservables”. In the next section we consider a case of “selection on observables”.
Example 2: Probit Selection on Observables
Suppose that we wish to know the mean and variance of 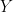 in the population. However, our sample of
in the population. However, our sample of 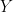 suffers from selection bias. In particular, there is some
suffers from selection bias. In particular, there is some 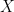 such that the probability of observing
such that the probability of observing 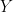 depends on
depends on 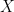 according to,
according to,
 ,
,
where  is some function with range
is some function with range ![[0,1] [0,1]](https://s0.wp.com/latex.php?latex=%5B0%2C1%5D&bg=ffffff&%23038;fg=333333&%23038;s=0) . Notice that, if
. Notice that, if 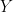 and
and 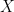 were independent, then the resulting sample distribution of
were independent, then the resulting sample distribution of 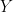 would be a random draw from the population (marginal distribution) of
would be a random draw from the population (marginal distribution) of 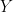 . Instead, we suppose
. Instead, we suppose 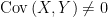 . For example,
. For example,
srand(2)
N = 10000
populationData = DataFrame(rand(MvNormal([0,0.],[1 .5;.5 1]),N)')
names!(populationData,[:X,:Y])
mean(array(populationData),1)
1x2 Array{Float64,2}:
-0.0281916 -0.022319
cov(array(populationData))
2x2 Array{Float64,2}:
0.98665 0.500912
0.500912 1.00195
In this simulated population, the estimated mean and variance of 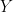 are -0.022 and 1.002, and the covariance between
are -0.022 and 1.002, and the covariance between 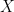 and
and 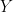 is 0.501. Now, suppose the probability that
is 0.501. Now, suppose the probability that  is observed is a probit regression of
is observed is a probit regression of 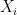 ,
,
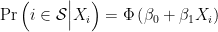 ,
,
where  is the CDF of the standard normal distribution. Letting
is the CDF of the standard normal distribution. Letting  indicate that
indicate that 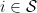 , we can generate the sample selection rule
, we can generate the sample selection rule 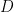 as,
as,
beta_0 = 0
beta_1 = 1
index = (beta_0 + beta_1*data[:X])
probability = cdf(Normal(0,1),index)
D = zeros(N)
for i=1:N
D[i] = rand(Bernoulli(probability[i]))
end
populationData[:D] = D
sampleData = populationData
sampleData[D.==0,:Y] = NA
The sample data has missing values in place of  if
if  . The estimated mean and variance of
. The estimated mean and variance of 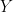 in the sample data are 0.275 (which is too large) and 0.862 (which is too small).
in the sample data are 0.275 (which is too large) and 0.862 (which is too small).
Correction 2: Inverse Probability Weighting
The reason for the biased estimates of the mean and variance of 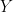 in Example 2 is sample selection on the observable
in Example 2 is sample selection on the observable 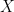 . In particular, certain values of
. In particular, certain values of 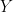 are over-represented due to their relationship with
are over-represented due to their relationship with 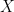 . Inverse probability weighting is a way to correct for the over-representation of certain types of individuals, where the “type” is captured by the probability of being included in the sample.
. Inverse probability weighting is a way to correct for the over-representation of certain types of individuals, where the “type” is captured by the probability of being included in the sample.
In the above simulation, conditional on appearing in the population, the probability that an individual of type  is included in the sample is 0.841. By contrast, the probability that an individual of type
is included in the sample is 0.841. By contrast, the probability that an individual of type  is included in the sample is 0.5, so type
is included in the sample is 0.5, so type 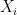 is over-represented by a factor of 0.841/0.5 = 1.682. If we could reduce the impact that type
is over-represented by a factor of 0.841/0.5 = 1.682. If we could reduce the impact that type  has in the computation of the mean and variance of
has in the computation of the mean and variance of 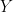 by a factor of 1.682, we would alter the balance of types in the sample to match the balance of types in the population. Inverse probability weighting generalizes this logic by weighting each individual’s impact by the inverse of the probability that this individual appears in the sample.
by a factor of 1.682, we would alter the balance of types in the sample to match the balance of types in the population. Inverse probability weighting generalizes this logic by weighting each individual’s impact by the inverse of the probability that this individual appears in the sample.
Before we can make the correct, we must first estimate the probability of sample inclusion. This can be done by fitting the probit regression above by least-squares. For this, we use the GLM package in Julia, which can be installed the usual way with the command Pkg.add(“GLM”).
using GLM
Probit = glm(D ~ X, sampleData, Binomial(), ProbitLink())
DataFrameRegressionModel{GeneralizedLinearModel,Float64}:
Coefficients:
Estimate Std.Error z value Pr(>|z|)
(Intercept) 0.114665 0.148809 0.770554 0.4410
X 1.14826 0.21813 5.26414 1e-6
estProb = predict(Probit)
weights = 1./estProb[D.==1]/sum(1./estProb[D.==1])
which are the inverse probability weights needed to match the sample distribution to the population distribution.
Now, we use the inverse probability weights to correct the mean and variance estimates of 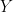 ,
,
correctedMean = sum(sampleData[D.==1,:Y].*weight)
-0.024566923132025013
correctedVariance = (N/(N-1))*sum((sampleData[D.==1,:Y]-correctedMean).^2.*weight)
1.0094029613131092
which are very close to the population values of -0.022319 and 1.00195. The logic here extends to the case of multivariate 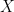 , as more coefficients are added to the Probit regression. The logic also extends to other functional forms of
, as more coefficients are added to the Probit regression. The logic also extends to other functional forms of  , for example, switching from Probit to Logit is achieved by replacing the ProbitLink() with LogitLink() in the glm() estimation above.
, for example, switching from Probit to Logit is achieved by replacing the ProbitLink() with LogitLink() in the glm() estimation above.
Example 3: Generalized Roy Model
For the final example of this tutorial, we consider a model which allows for rich, realistic economic behavior. In words, the Generalized Roy Model is the economic representation of a world in which each individual must choose between two options, where each option has its own benefits, and one of the options costs more than the other. In math notation, the first alternative, denoted  , relates the outcomes
, relates the outcomes  to the individual’s observable characteristics,
to the individual’s observable characteristics, 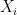 , by,
, by,
 .
.
Similarly, the second alternative, denoted  , relates
, relates  to
to 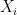 , by,
, by,
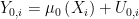 .
.
The value of  that appears in our sample is thus given by,
that appears in our sample is thus given by,
 .
.
Finally, the value of  is chosen by individual
is chosen by individual  according to,
according to,
 if
if 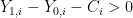 ,
,
where  is the cost of choosing the alternative
is the cost of choosing the alternative  and is given by,
and is given by,
 ,
,
where 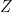 contains additional characteristics of
contains additional characteristics of  that are not included in
that are not included in 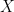 .
.
We assume that the data only contains  ; it does not contain the variables
; it does not contain the variables  or the functions
or the functions  . Assuming that the three
. Assuming that the three 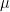 functions follow the linear form and that the unobservables
functions follow the linear form and that the unobservables 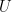 are independent and Normally distributed, we can simulate the data generating process as,
are independent and Normally distributed, we can simulate the data generating process as,
srand(2)
N = 1000
sampleData = DataFrame(rand(MvNormal([0,0.],[1 .5; .5 1]),N)')
names!(sampleData,[:X,:Z])
U1 = rand(Normal(0,.5),N)
U0 = rand(Normal(0,.7),N)
UC = rand(Normal(0,.9),N)
betas1 = [0,1]
betas0 = [.3,.2]
betasC = [.1,.1,.1]
Y1 = betas1[1] + betas1[2].*sampleData[:X] + U1
Y0 = betas0[1] + betas0[2].*sampleData[:X] + U0
C = betasC[1] + betasC[2].*sampleData[:X] + betasC[3].*sampleData[:Z] + UC
D = Y1-Y0-C.>0
Y = D.*Y1 + (1-D).*Y0
sampleData[:D] = D
sampleData[:Y] = Y
In this simulation, about 38% of individuals choose the alternative  . About 10% of individuals choose
. About 10% of individuals choose  even though they receive greater benefits under
even though they receive greater benefits under  due to the high cost
due to the high cost  associated with
associated with  .
.
Solution 3: Heckman Correction for Generalized Roy Model
The identification of this model is attributable to Heckman and Honore (1990). Estimation proceeds in steps. The first step is to notice that the left- and right-hand terms in the following moment equation motivate a Probit regression:
![\mathbb{E}\left[D_i\Big|X_i,Z_i\right]=\Pr\left(\mu_D\left(X_i,Z_i\right)>U_{D,i}\Big|X_i,Z_i\right)=\Phi\left(\mu_D\left(X_i,Z_i\right)\right) \mathbb{E}\left[D_i\Big|X_i,Z_i\right]=\Pr\left(\mu_D\left(X_i,Z_i\right)>U_{D,i}\Big|X_i,Z_i\right)=\Phi\left(\mu_D\left(X_i,Z_i\right)\right)](https://s0.wp.com/latex.php?latex=%5Cmathbb%7BE%7D%5Cleft%5BD_i%5CBig%7CX_i%2CZ_i%5Cright%5D%3D%5CPr%5Cleft%28%5Cmu_D%5Cleft%28X_i%2CZ_i%5Cright%29%3EU_%7BD%2Ci%7D%5CBig%7CX_i%2CZ_i%5Cright%29%3D%5CPhi%5Cleft%28%5Cmu_D%5Cleft%28X_i%2CZ_i%5Cright%29%5Cright%29&bg=ffffff&%23038;fg=333333&%23038;s=0) ,
,
where 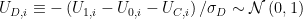 is the negative of the total error term arising in the equation that determines
is the negative of the total error term arising in the equation that determines  above,
above,  , and,
, and,
![\mu_D\left(X,Z\right) \equiv \left([1,X]\beta_1 -[1,X]\beta_0 -[1,X,Z]\beta_C\right)/\sigma_D \equiv [1,X,Z]\beta_D \mu_D\left(X,Z\right) \equiv \left([1,X]\beta_1 -[1,X]\beta_0 -[1,X,Z]\beta_C\right)/\sigma_D \equiv [1,X,Z]\beta_D](https://s0.wp.com/latex.php?latex=%5Cmu_D%5Cleft%28X%2CZ%5Cright%29+%5Cequiv+%5Cleft%28%5B1%2CX%5D%5Cbeta_1+-%5B1%2CX%5D%5Cbeta_0+-%5B1%2CX%2CZ%5D%5Cbeta_C%5Cright%29%2F%5Csigma_D+%5Cequiv+%5B1%2CX%2CZ%5D%5Cbeta_D&bg=ffffff&%23038;fg=333333&%23038;s=0) ,
,
In the simulation above, ![\beta_D = [-.4,.7,-.1]/\sqrt{.5+.7+.9}\approx[-0.276,0.483,-0.069] \beta_D = [-.4,.7,-.1]/\sqrt{.5+.7+.9}\approx[-0.276,0.483,-0.069]](https://s0.wp.com/latex.php?latex=%5Cbeta_D+%3D+%5B-.4%2C.7%2C-.1%5D%2F%5Csqrt%7B.5%2B.7%2B.9%7D%5Capprox%5B-0.276%2C0.483%2C-0.069%5D&bg=ffffff&%23038;fg=333333&%23038;s=0) . We can estimate
. We can estimate  from the Probit regression of
from the Probit regression of 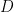 on
on 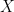 and
and 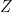 .
.
betasD = coef(glm(D~X+Z,sampleData,Binomial(),ProbitLink()))
3-element Array{Float64,1}:
-0.299096
0.59392
-0.103155
Next, notice that,
![\mathbb{E}\left[Y_i\Big|D_i=1,X_i,Z_i\right] =[1,X_i]\beta_1+\mathbb{E}\left[U_{1,i}\Big|D_i=1,X_i,Z_i\right] \mathbb{E}\left[Y_i\Big|D_i=1,X_i,Z_i\right] =[1,X_i]\beta_1+\mathbb{E}\left[U_{1,i}\Big|D_i=1,X_i,Z_i\right]](https://s0.wp.com/latex.php?latex=%5Cmathbb%7BE%7D%5Cleft%5BY_i%5CBig%7CD_i%3D1%2CX_i%2CZ_i%5Cright%5D+%3D%5B1%2CX_i%5D%5Cbeta_1%2B%5Cmathbb%7BE%7D%5Cleft%5BU_%7B1%2Ci%7D%5CBig%7CD_i%3D1%2CX_i%2CZ_i%5Cright%5D&bg=ffffff&%23038;fg=333333&%23038;s=0) ,
,
where,
![\mathbb{E}\left[U_{1,i}\Big|D_i=1,X_i,Z_i\right] =\mathbb{E}\left[U_{1,i}\Big|\mu_D\left(X_i,Z_i\right)>U_{i,D},X_i,Z_i\right]=\rho_1 \frac{-\phi\left(\mu_D\left(X_i,Z_i\right)\right)}{\Phi\left(\mu_D\left(X_i,Z_i\right)\right)} \mathbb{E}\left[U_{1,i}\Big|D_i=1,X_i,Z_i\right] =\mathbb{E}\left[U_{1,i}\Big|\mu_D\left(X_i,Z_i\right)>U_{i,D},X_i,Z_i\right]=\rho_1 \frac{-\phi\left(\mu_D\left(X_i,Z_i\right)\right)}{\Phi\left(\mu_D\left(X_i,Z_i\right)\right)}](https://s0.wp.com/latex.php?latex=%5Cmathbb%7BE%7D%5Cleft%5BU_%7B1%2Ci%7D%5CBig%7CD_i%3D1%2CX_i%2CZ_i%5Cright%5D+%3D%5Cmathbb%7BE%7D%5Cleft%5BU_%7B1%2Ci%7D%5CBig%7C%5Cmu_D%5Cleft%28X_i%2CZ_i%5Cright%29%3EU_%7Bi%2CD%7D%2CX_i%2CZ_i%5Cright%5D%3D%5Crho_1+%5Cfrac%7B-%5Cphi%5Cleft%28%5Cmu_D%5Cleft%28X_i%2CZ_i%5Cright%29%5Cright%29%7D%7B%5CPhi%5Cleft%28%5Cmu_D%5Cleft%28X_i%2CZ_i%5Cright%29%5Cright%29%7D&bg=ffffff&%23038;fg=333333&%23038;s=0) ,
,
which is the inverse Mills ratio again, where  . Substituting in the estimate for
. Substituting in the estimate for  , we consistently estimate
, we consistently estimate 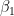 :
:
fittedVals = hcat(ones(N),array(sampleData[[:X,:Z]]))*betasD
sampleData[:invMills1] = -pdf(Normal(0,1),fittedVals)./cdf(Normal(0,1),fittedVals)
correctedBetas1 = linreg(array(sampleData[D,[:X,:invMills1]]),vec(array(sampleData[D,[:Y]])))
3-element Array{Float64,1}:
0.0653299
0.973568
-0.135445
To see how well the correction has performed, compare these estimates to the uncorrected estimates of 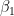 ,
,
biasedBetas1 = linreg(array(sampleData[D,[:X]]),vec(array(sampleData[D,[:Y]])))
2-element Array{Float64,1}:
0.202169
0.927317
Similar logic allows us to estimate  :
:
sampleData[:invMills0] = pdf(Normal(0,1),fittedVals)./(1-cdf(Normal(0,1),fittedVals))
correctedBetas0 = linreg(array(sampleData[D.==0,[:X,:invMills0]]),vec(array(sampleData[D.==0,[:Y]])))
3-element Array{Float64,1}:
0.340621
0.207068
0.323793
biasedBetas0 = linreg(array(sampleData[D.==0,[:X]]),vec(array(sampleData[D.==0,[:Y]])))
2-element Array{Float64,1}:
0.548451
0.295698
In summary, we can consistently estimate the benefits associated with each of two alternative choices, even though we only observe each individual in one of the alternatives, subject to heavy selection bias, by extending the logic introduced by Heckman (1979).
Bradley J. Setzler